Getting Started With CI/CD for Data Science Pipelines
Our Approach to Using CI/CD For Data Science
When it comes to training and scoring machine learning models, there are trade-offs to using your local machine vs. CI. Our approach is flexible to allow users to do both.
-
When executing training or scoring runs on your local machine, users will be able to:
- Execute training or scoring code using local resources and python builds specific to the machine
- Log, view, and retrieve experiments in your preferred experiment tracker
- Upload and download model artifacts to your preferred model/package registry
- Commit changes to the repository that will not trigger a CI pipeline
-
When executing training or scoring runs remotely using GitLab CI/CD, users will be able to:
- Select CPU or GPU compute instance based on the needs of the model
- Automatically detect and rebuild the model image based on changes to Dockerfile or requirements.txt
- Log, view, and retrieve experiments in your preferred experiment tracker
- Upload and download model artifacts directly to the GitLab package registry
- Automatically report model metrics and performance in the merge request for review by others.
- Commit changes to the repository and automatically trigger training CI pipelines based on a specific commit message
- Training CI pipelines only execute with the following commit message:
train <path/to/notebook/your_notebook.ipynb> - Allows the pipeline to execute just the desired notebook
- Training CI pipelines only execute with the following commit message:
- Commit changes to the repository and automatically trigger the scoring CI pipeline based on a specific commit message
- Scoring CI pipeline only executes with the following commit message:
score <path/to/notebook/your_notebook.ipynb>
- Scoring CI pipeline only executes with the following commit message:
- Allow training and scoring CI pipelines to run at set dates and times using Scheduled pipelines
- Log pipeline results to Project wiki
- Use GitLab for Slack integration to monitor pipeline status
Advantages of Using CI for Training Data Science Models
- Reproducibility
- Automation
- Speed
- Logging results directly to Merge Request and Project Wiki
- Scalable GPU and CPU resources
- Scheduling
- Slack notifications for monitoring CI pipelines
Getting Started
This section covers, in detail, the mechanisms behind how these pipelines is created and configured. If you are just interested in getting your data science CI training pipeline up and running, skip directly to the Model Training Step-by-Step Instructions
If you are just interested in getting your data science CI scoring pipeline up and running, skip directly to the Scoring and Productionalization Step-by-Step Instructions
Key Repository Files
Within our public GitLab Data Science CI Example repository are the following key files:
- .gitlab-ci.yml: This is the CI/CD configuration file that define the jobs that define the jobs that will be run in each pipeline. The actual pipelines are pulled from the CI/CD Component Catelog, with only the variables that need specified by the user set in this .yml.
- Dockerfile: Instructions for creating the docker image. Here we are using python 3.9 running on Ubuntu 22.04 with CUDA drivers for GPU
- requirements.txt: The python packages to install in the docker container
- training_config.yaml: Configuration for training notebook
- scoring_config.yaml: Configuration for scoring notebook
- notebooks/training_example.ipynb: training notebook used for this example
- notebooks/scoring_example.ipynb: scoring code productionalization notebook used for this example
- xgb_model.json: The saved model from training that will be used for scoring (Note: in future iterations this will be pulled directly from the Model Registry)
Model Training with CI/CD
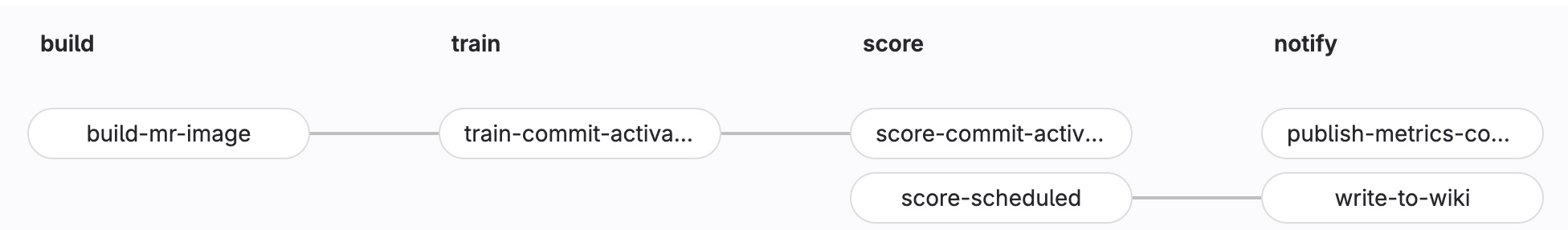
Training Pipeline
- Build
- build-ds-image: activated whenever changes are made to the Dockerfile or requirements.txt files. This will rebuild the image used to train the model
- Train
- train-commit-activated: To execute a training pipeline. Activated by using the
train <path/to/notebook/your_notebook.ipynb>commit message
- train-commit-activated: To execute a training pipeline. Activated by using the
- Notify (optional)
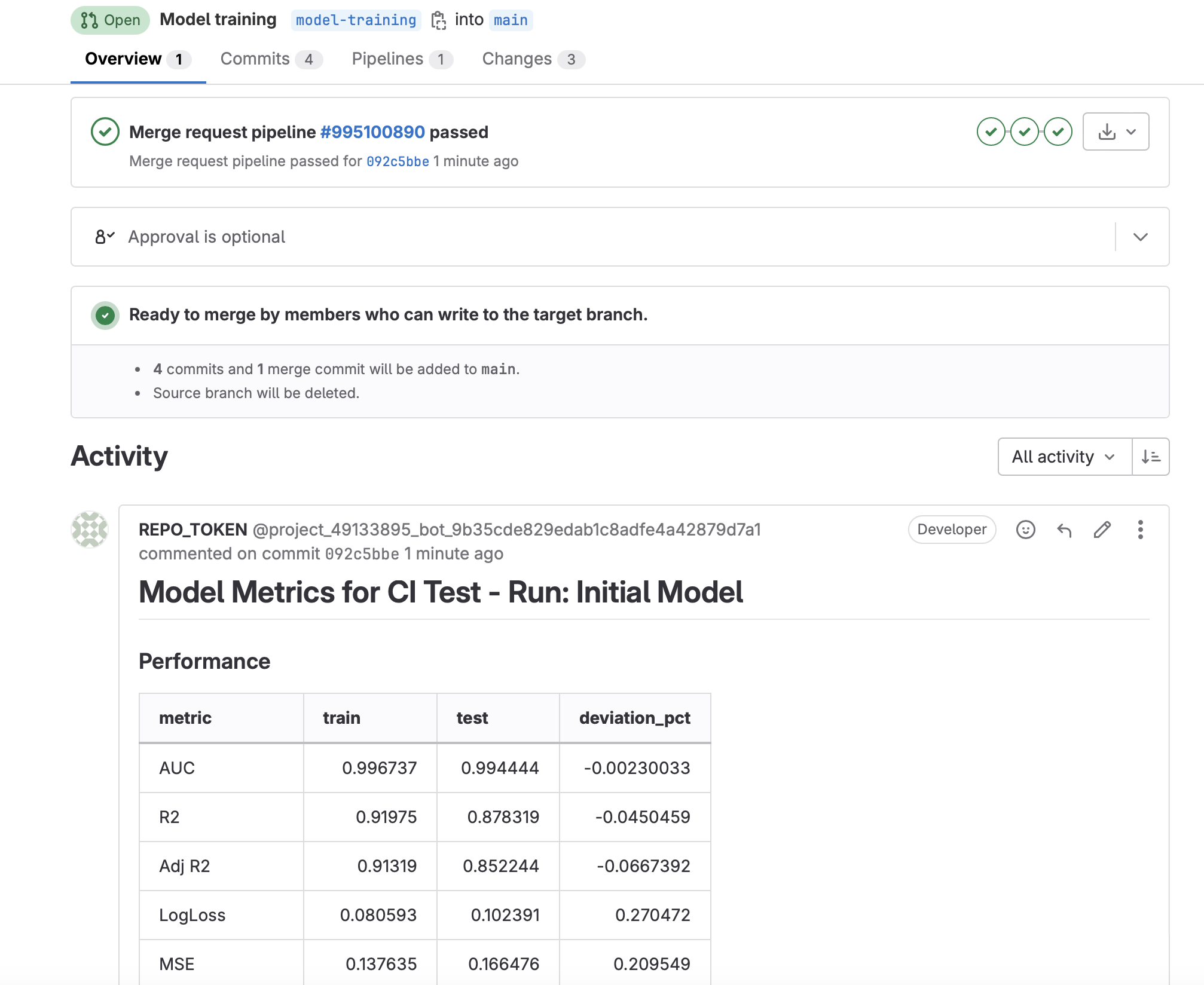
- publish-metrics-comment: Write model metrics as a comment to the merge request. This is executed after a training or scoring run is performed via commit message.
Training Setup
Let’s take a detailed look at the repository (Code -> Repository):
- In the notebooks directory, open training_example.ipynb. We do not need to change anything in here, but note the first cell of the notebook. Papermill will be used to execute this notebook in the CI pipeline and this cell has been tagged as
parametersto allow Papermill to change these values. There are 3 variables with preset values:is_local_development = True: The default assumption is that the notebook is being run locally. The CI pipeline will modify this value toFalseautomatically so the notebook can be optimized to run in CItree_method = 'auto': The default value for xgboost. The CI pipeline changes this value togpu_histwhen using a GPU runner in CI to take advantage certain performance enhancements. The assumption that is the model will train via CPU when run locally due to lack of a compatible local GPU.notebook_dir = 'notebooks': The directory of the training notebooks. This value is the same locally as it is the CI pipeline.
- To see exactly how the CI pipeline changes these values, let’s view .gitlab-ci.yml. Because we are using a component from the CI/CD catalog, we actually want to look at the Data Science ML Component Pipeline. In this file, search for
train-commit-activated. This is the most important stage in the CI pipeline. A few things to note:image: $CONTAINER_IMAGE: The training job will use the container created in the build job (as defined bybuild-ds-image), using the Dockerfile and requirements.txt files in the repository.tags: Determines which runner to use. We want to be able to change the runner based on the project needs. This gets specified byTRAIN_RUNNERin .gitlab-ci.yml located back in our project repositoryscript: … -papermill -p is_local_development False -p tree_method 'gpu_hist' $notebookName -: Tells Papermill to override the variable values defined in the first cell of the notebook with the values shown when utilizing a the GPU runner.
- Finally, let’s look at the training_config.yaml. Here we can configure certain variables for training our model:
outcome: Our outcome/target/dv variable. The example notebook is using the breast cancer dataset from scikit-learn and the outcome field in that dataset is namedtargetoptunaconfigurations: The example notebook runs an xgboost model with Optuna. There are a lot of customizations possible with this setup, but to keep it simple we have only included:n_trials: Number of trails to run in the Optuna studymodel_file_name: The output name of the model file
mlflow: There are a few configurations you can make here if you like, but the defaults will also work fine for this exampleexperiment_name: Name of your MLFlow Experimentrun_name: ID or name of the MLFlow Experiment Run
Model Training Step-by-Step Instructions
- Fork the public GitLab Data Science CI Example repository. Forking will allow you to further customize the code to meet your own needs.
- Optional (but recommended) Configurations:
- Experiment Tracker: This will allow you to log your experiments in the native Experiment Tracker (or MLFlow instance) and log the model artifacts to the package registry.
- Review the MLflow Client Compatibility Instructions to set up the
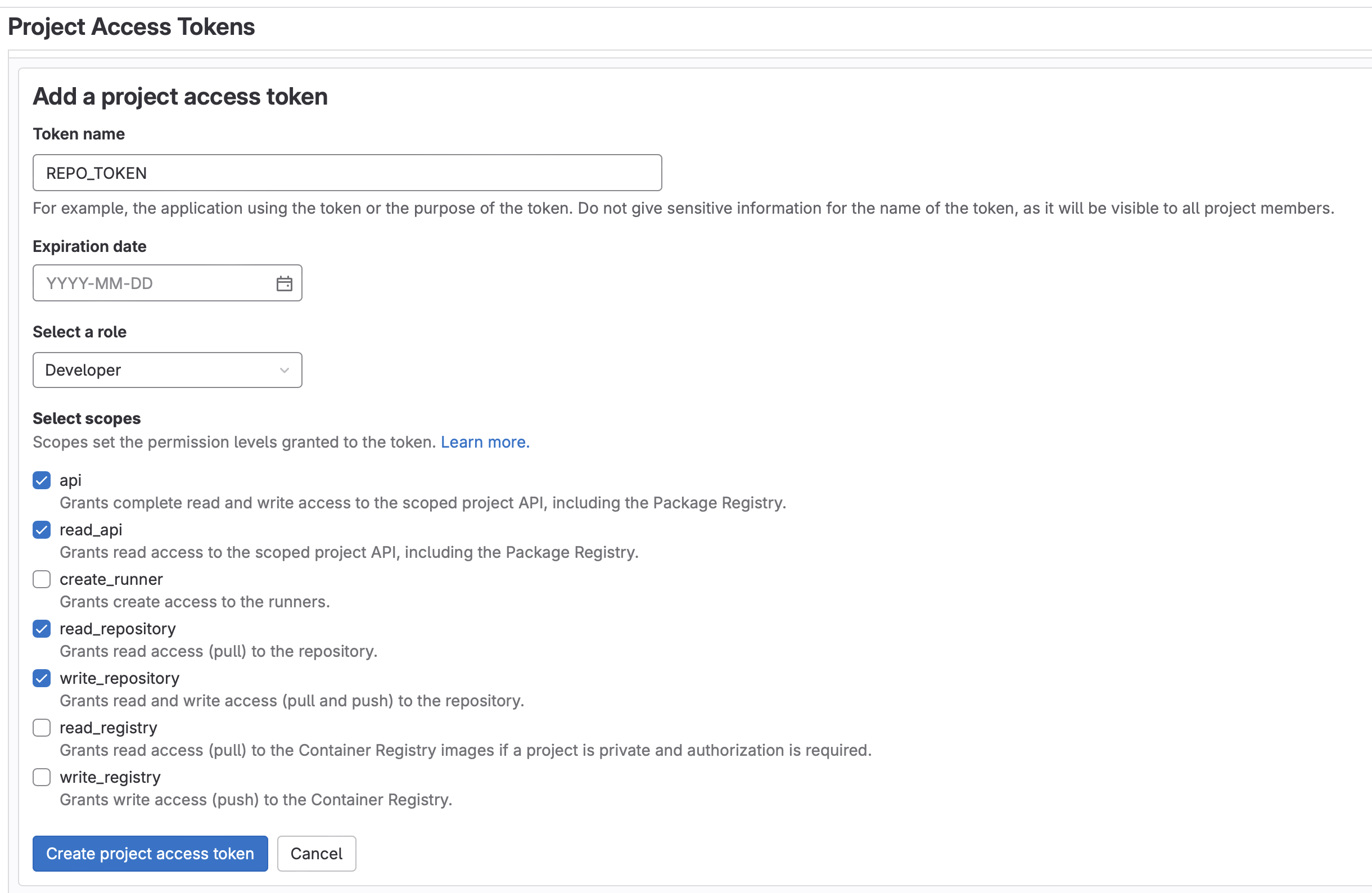
MLFLOW_TRACKING_URIandMLFLOW_TRACKING_TOKENCI/CI in your project. - Create a project access token (Settings -> Access Tokens -> Add New Token) named
REPO_TOKENwith aDeveloperrole and the following scopes:api, read_api, read_repository, write_repository. Be sure to copy this token.- Note: Enabling group access tokens is a not available for SaaS Free accounts. If using a Free account, you will need to fork the project into a personal (instead of a group) namespace
- Note: Enabling group access tokens is a not available for SaaS Free accounts. If using a Free account, you will need to fork the project into a personal (instead of a group) namespace
- Create the following new CI Variables (Settings -> CI/CD -> Variables -> Add New Variable):
MLFLOW_TRACKING_TOKEN: For the value, enter the project access token value created above.MLFLOW_TRACKING_URI: For the value, use the GitLab API MLFlow endpoint as outlined in the MLFlow instructions above. It should look something like:https://gitlab.com/api/v4/projects/<your_project_id>/ml/mlflow. Project ID can be found in Settings -> General- Note: For all varibles, untick the “Protect Variable” flag to enable experiment tracking on unprotected branches. Tick “Mask variable” to prevent the value from showing in the logs.
- Review the MLflow Client Compatibility Instructions to set up the
- Write Model Metrics to Merge Request
- Create the following new CI/CD Variable (Settings -> CI/CD -> Variables -> Add New Variable):
REPO_TOKEN: For the value, enter the project access token value created above.- Note: Untick the “Protect Variable” flag to enable experiment tracking on unprotected branches. Tick “Mask variable” to prevent the value from showing in the logs.
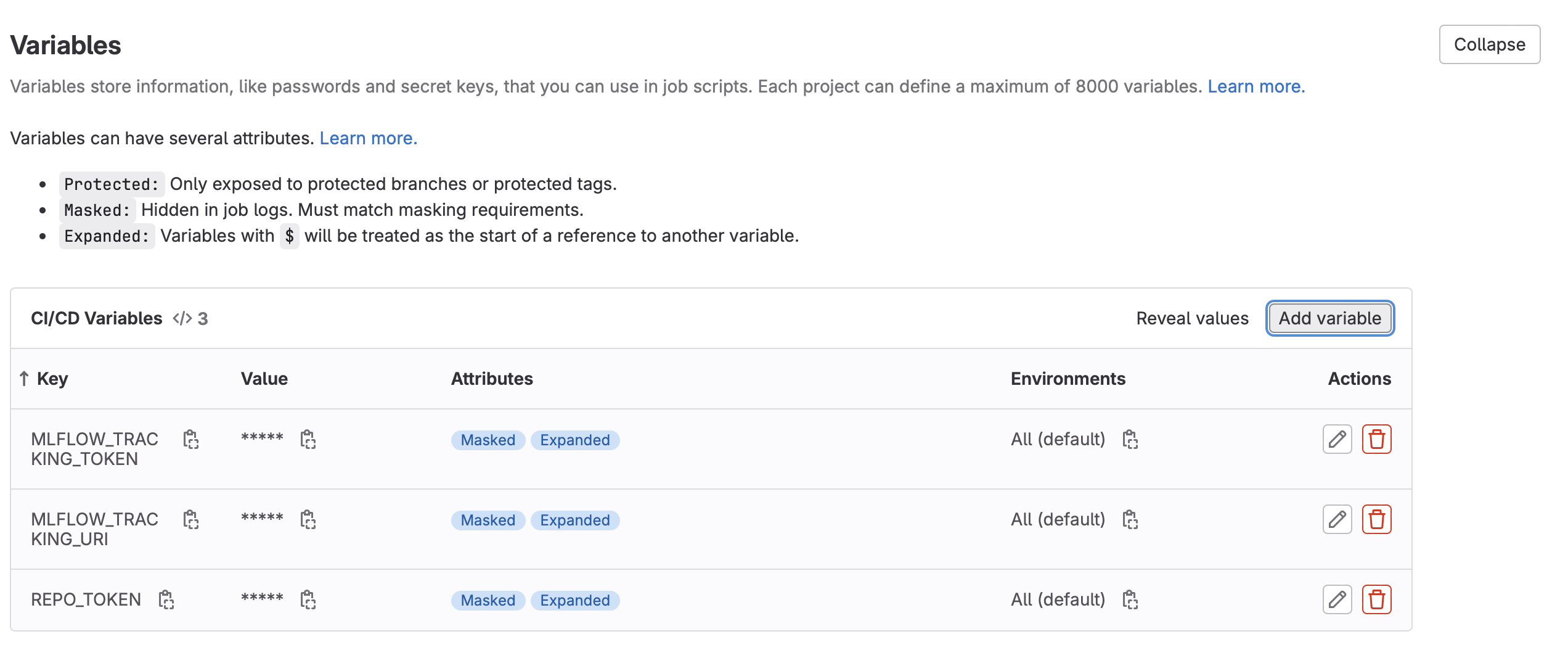
- Create the following new CI/CD Variable (Settings -> CI/CD -> Variables -> Add New Variable):
- Experiment Tracker: This will allow you to log your experiments in the native Experiment Tracker (or MLFlow instance) and log the model artifacts to the package registry.
- Now, let’s make some changes to activate our training pipeline:
- Create a new branch (Code -> Branches -> New Branch)
-
- First let’s make sure the
build-ds-imagewill get triggered, which will build the container our model will run in. This job of the pipeline is only activated when changes are detected in Dockerfile or requirements.txt. So let’s make a change:- Edit Dockerfile, replacing the maintainer value with your GitLab handle and commit the change to your branch.
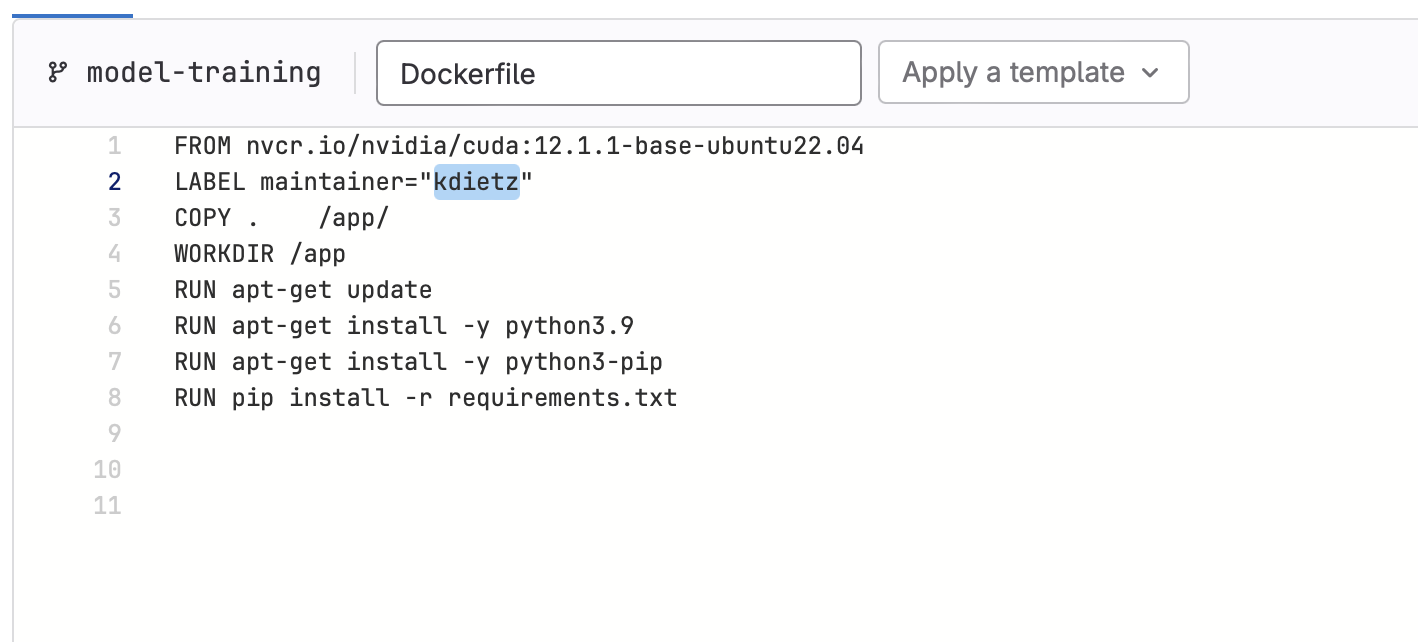
- Edit Dockerfile, replacing the maintainer value with your GitLab handle and commit the change to your branch.
- Configure your runners:
- GPU runners are available for Premium and Ultimate users. If enabled, edit
.gitlab-ci.ymland change the value ofTRAIN_RUNNERto a GPU runner (i.e.saas-linux-medium-amd64-gpu-standard). - The default value,
saas-linux-small-amd64, will work for all account types.
- GPU runners are available for Premium and Ultimate users. If enabled, edit
- Edit training_config.yaml
- Change
n_trialsto a new value between10and20. - For the commit message enter
train notebooks/training_example.ipynb. This will tell the GitLab that you want to execute the training CI pipeline on the training_example.ipynb notebook found in the notebooks directory. Commit the change.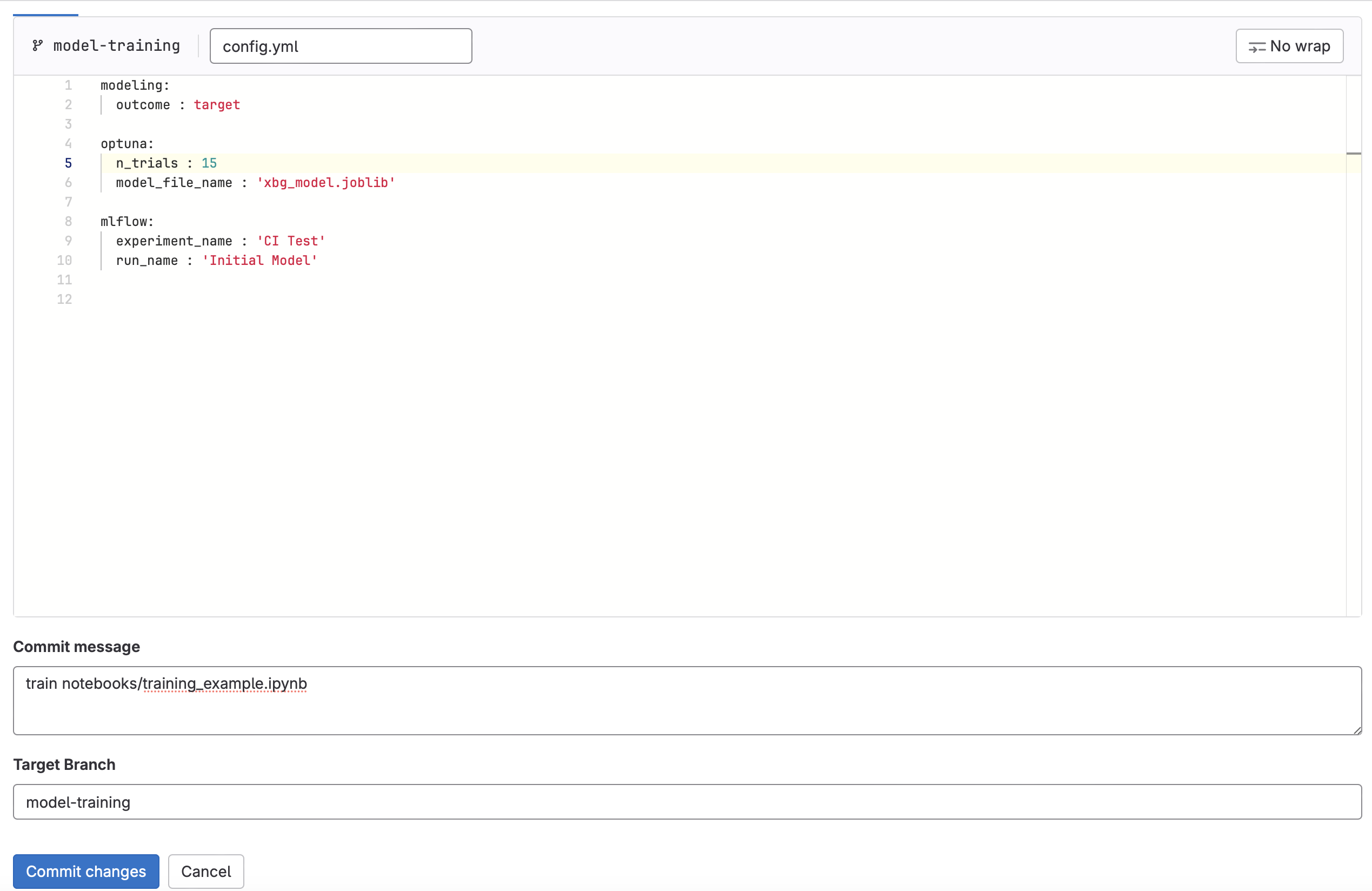
- Change
- Click “Create merge request”. Make sure you are merging into your local fork and click “Create merge request” once again. This should activate the training CI pipeline for the newly created MR.
- Click on “Pipelines” and you should see the training pipeline running. Click into the pipeline to see which stage the pipeline is in.
- Note: If you did not set up the step above “Write Model Metrics to Merge Request”, then the
publish-metrics-commentjob will fail. The pipeline will still pass with warnings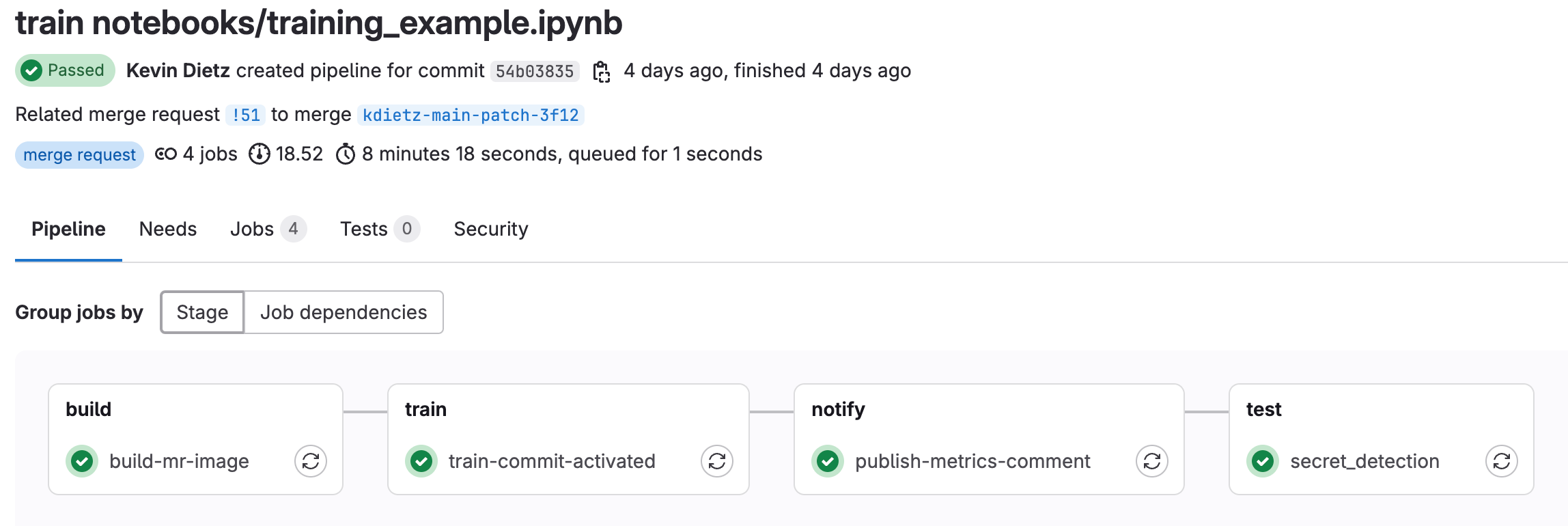
- Note: If you did not set up the step above “Write Model Metrics to Merge Request”, then the
- Once the pipeline has finished, you will see a new comment posted on the merge request that contains some model metrics from the run (assuming you set up Write Model Metrics to Merge Request).
-
- Now let’s look at the experiment run we just completed with our CI pipeline (Analyze -> Model Experiments)
- Click on your experiment name.
- You should see a new run logged from the CI Pipeline. Click into that run.
- Run details are displayed, including a link to the CI job, the merge request, various parameters and metrics, and model artifacts.
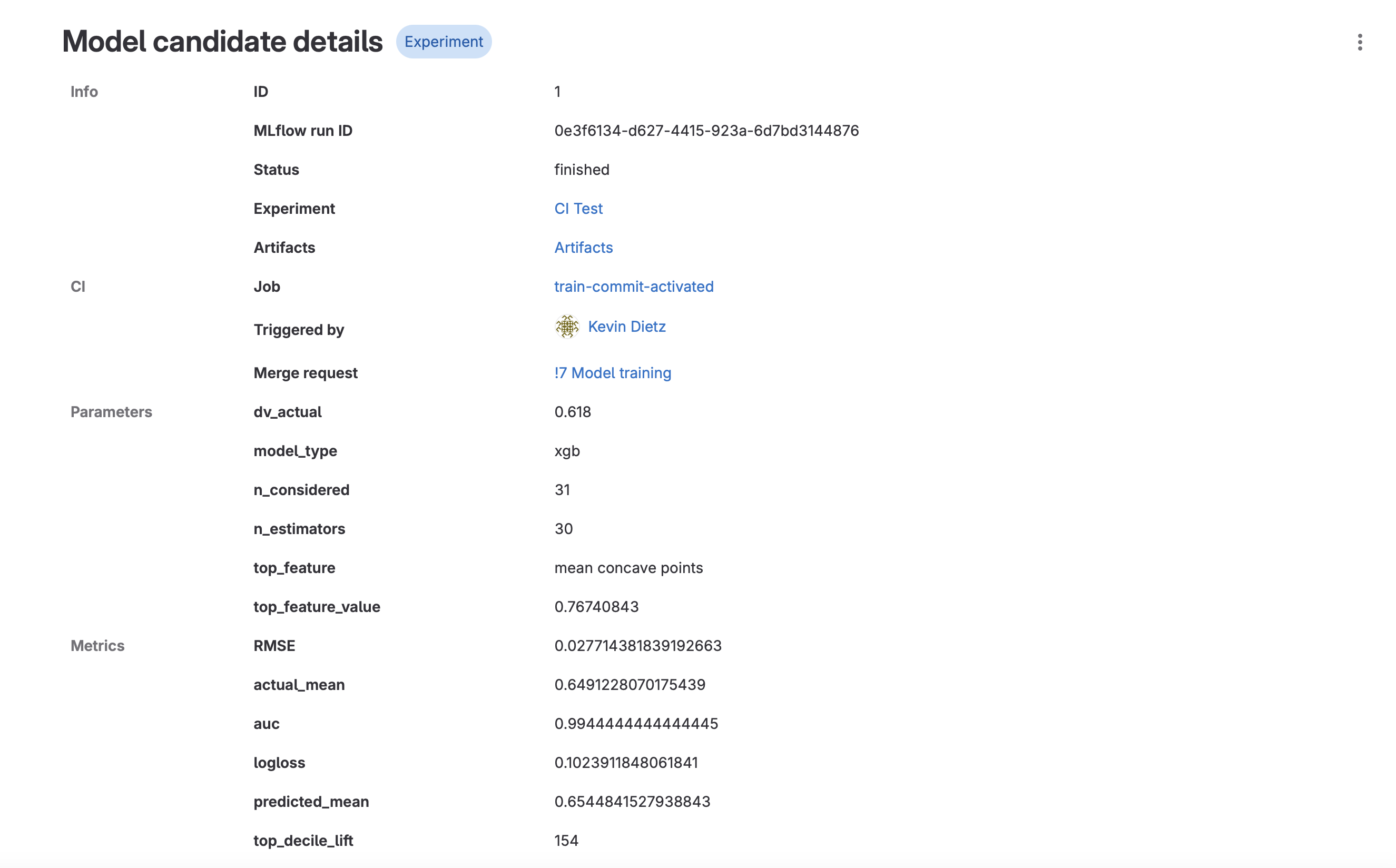
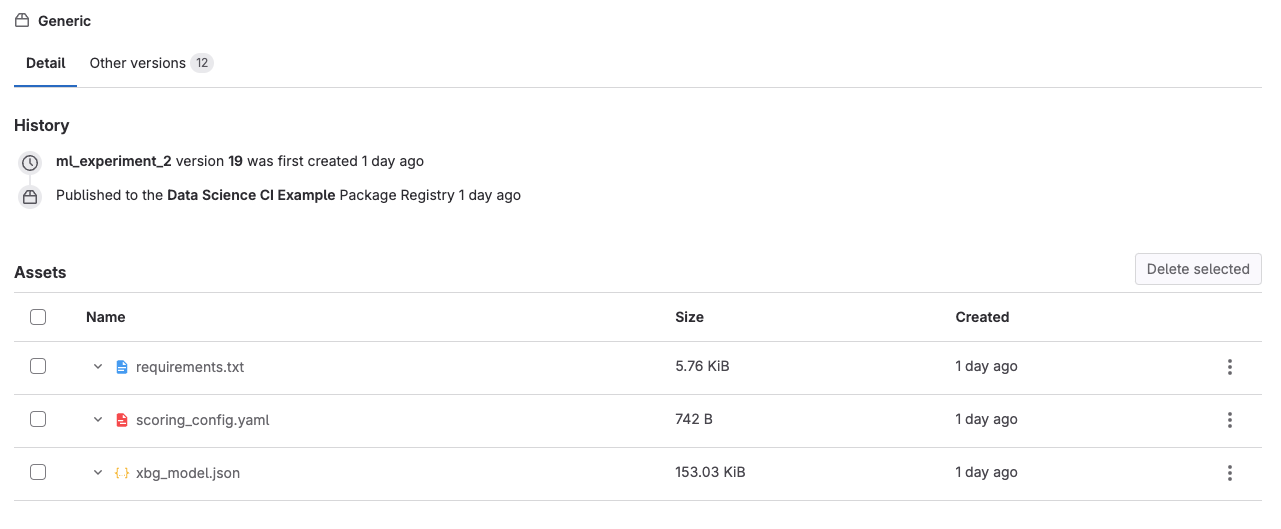
- Click on “Artifacts”. This will take you to the Package Registry, where all the artifacts associated with that particular model run are stored. You should see the .json model file, a .yaml configuration file, and a requirements.txt. These can be used later to deploy your model.
- Finally, let’s look at the container that was used to train the model (Deploy -> Container Registry)
- This container will be used in subsequent runs of the model and will only get rebuilt when Dockerfile or requirements.txt are modified.
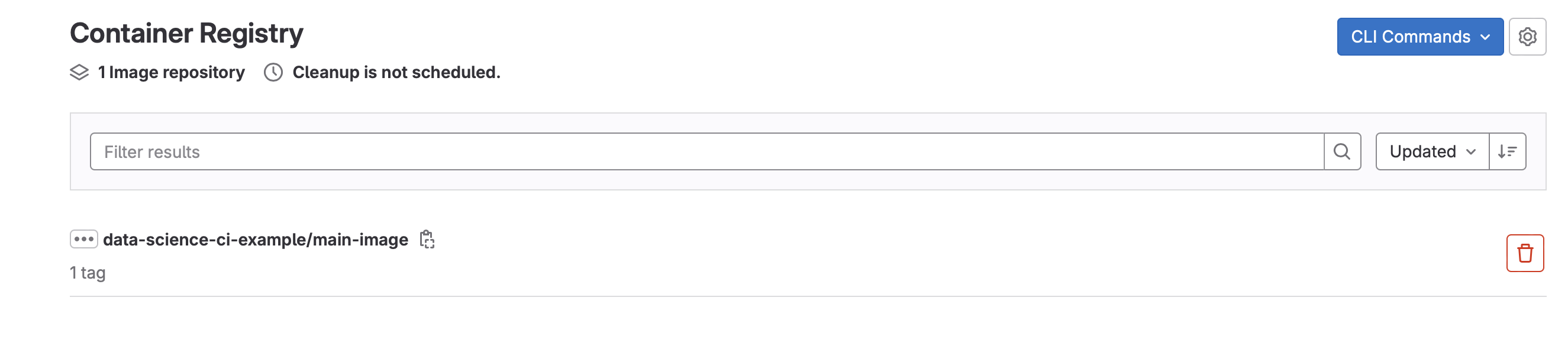
- This container will be used in subsequent runs of the model and will only get rebuilt when Dockerfile or requirements.txt are modified.
Scoring and Productionalization with CI/CD
Scoring Pipeline
- Build
- build-ds-image: activated whenever changes are added to the Dockerfile or requirements.txt files. This will rebuild the image used to score the model. Ideally, this should be the same image that was used to train the model.
- Score
- score-commit-activated: To manually execute a scoring pipeline. Activated by using
score <path/to/notebook/your_notebook.ipynb>in the commit message - score-scheduled: To execute a scoring pipeline based on a defined schedule using Scheduled pipelines
- score-commit-activated: To manually execute a scoring pipeline. Activated by using
- Notify (Optional)
- publish-metrics-comment: Write model performance metrics as a comment on the merge request. This is executed after a scoring run is performed via commit message.
- write-to-wiki: Write model performance metrics and job details to the project wiki. The CI configuration it set up to only execute for scheduled jobs that use the
score-scheduledjob

Productionalization Setup
Let’s take a detailed look at the repository (Code -> Repository):
- In the notebooks directory, open scoring_example.ipynb. We do not need to change anything in here, but note the first cell of the notebook. Papermill will be used to execute this notebook in the CI pipeline and this cell has been tagged as
parametersto allow Papermill to change these values. There are 2 variables with preset values:is_local_development = True: The default assumption is that the notebook is being run locally. The CI pipeline will modify this value toFalseautomatically so the notebook can be optimized to run in CInotebook_dir = 'notebooks': The directory of the scoring notebook (can be the same directory as the training notebooks). This value is the same locally as it is the CI pipeline.
- To see exactly how the CI pipeline changes these values, now let’s view .gitlab-ci.yml. Because we are using a component from the CI/CD catelog, we actually want to look at the Data Science ML Component Pipeline. In this file, search for
score-commit-activated. A few things to note:- This job will manually trigger when
score <path/to/notebook/your_notebook.ipynb>is passed in the commit message. image: $CONTAINER_IMAGE: The scoring job will use the container created in the build job (as defined bybuild-ds-image), using the Dockerfile and requirements.txt files in the repository.tags: Determines which runner to use. We want to be able to change the runner based on the project needs. This gets specified bySCORE_RUNNERin .gitlab-ci.yml located back in our project repositoryscript: … -papermill -p is_local_development False $notebookName -: Tells Papermill to override the variable values defined in the first cell of the notebook with the values shown when running in CI.
- This job will manually trigger when
- There is also a
score-scheduledjob.- This will trigger the scoring notebook at a set time, using Scheduled pipelines
- This job will also trigger the
write-to-wikijob, which will publish model metrics to the project wiki
- Finally, let’s look at the scoring_config.yaml. Here we can configure certain variables for training our model:
- model_file: The model file created during training that will be used to score the model. This could also be pulled directly from the Model Registry, but for simplicity, we are including it directly in the repository
- fields: List of the model fields. This is useful if the model is using only a subset of fields in a dataframe. In this example, all the fields in the dataframe are being used.
- decile_cuts: Sometimes it’s useful to include decile cuts from the validation dataset used during training. In this example, we included quintile cuts to illustrate how they can be used.
Scoring and Productionalization Step-by-Step Instructions
- Optional (but recommended) Wiki Configurations:
- This allows you to log your scheduled runs to the GitLab Project Wiki.
- Create a new CI/CD Variable (Settings -> CI/CD -> Variables -> Add New Variable):
API_ENDPOINT: For the value, use the GitLab API endpoint (will be similar to theMLFLOW_TRACKING_URIset up during training) using the following format:https://gitlab.com/api/v4/projects/<your_project_id>. Project ID can be found in Settings -> General.- Note: Untick the “Protect Variable” flag to enable experiment tracking on unprotected branches
- Configure your runner:
- GPU runners are available for Premium and Ultimate users. If enabled, edit
.gitlab-ci.ymland change the value ofSCORE_RUNNERto a GPU runner (i.e.saas-linux-medium-amd64-gpu-standard). - The default value,
saas-linux-small-amd64, will work for all account types.
- GPU runners are available for Premium and Ultimate users. If enabled, edit
- Now lets score our model using CI/CD
- You can either create a new branch of the repository or use the same one as used above for training the model.
- Make a change to
notebooks/scoring_example.ipynbso that we have something to commit. This could be as simple as adding a line to one of the cells. - For the commit message enter
score notebooks/scoring_example.ipynb. This will tell the GitLab that you want to execute the `score-commit-activated`` CI pipeline on the scoring_example.ipynb notebook found in the notebooks directory. Commit the change. - Click “Create merge request”. Make sure you are merging into your local fork and click “Create merge request” once again. This should activate the scoring CI pipeline for the newly created MR.
- Click on “Pipelines” and you should see the scoring pipeline running. Click into the pipeline to see which stage the pipeline is in.
- Note: If you did not set up the step above “Write Model Metrics to Merge Request”, then the
publish-metrics-commentjob will fail. The pipeline will still pass with warnings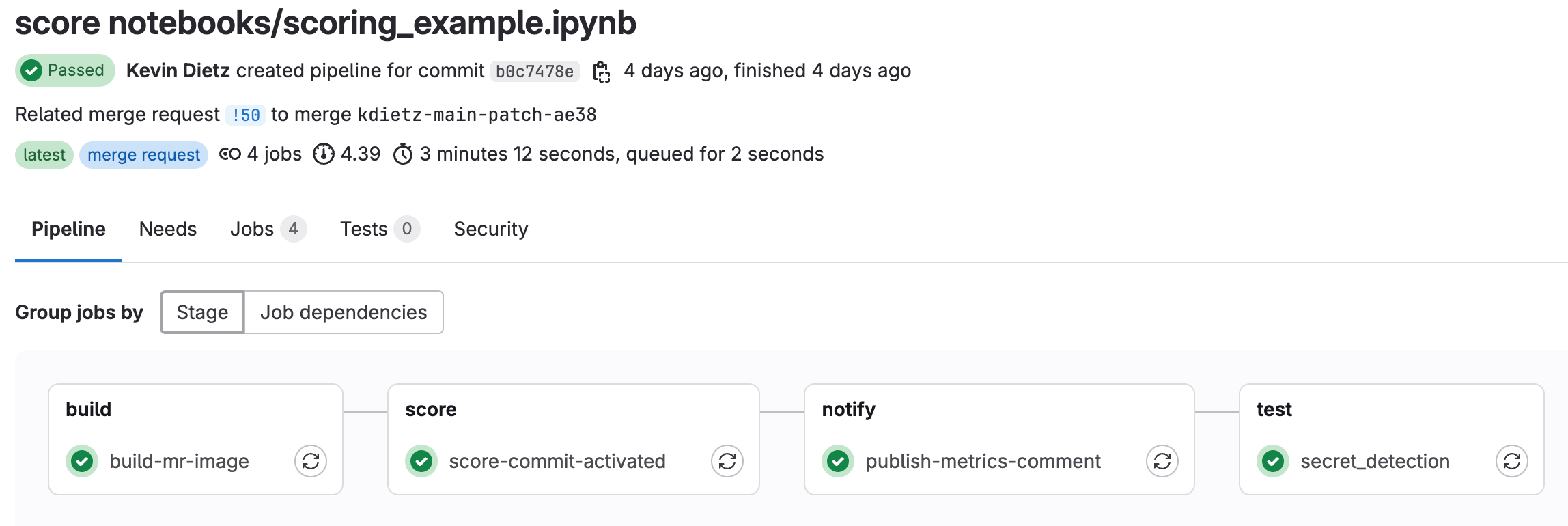
- Once the pipeline has finished, you will see a new comment posted on the merge request that contains some model metrics from the run (assuming you set up Write Model Metrics to Merge Request). This is the same process you would have seen during training, except it is now writting out descriptives about the scored dataset.
- We can also set the model to score at a defined time using Pipeline Schedules
- Nagivate to Build -> Pipeline schedules -> New schedule
- Give your pipeline a description
- Set when you want it to run. In the example below, the pipeline is scheduled to run every day at noon.
- Target branch:
mainwill work for this example. (You could also set it to the branch are you are currently working in.) - Variables: We only need to setup one variable,
SCORING_NOTEBOOKwith the location of the notebook we want to schedulenotebooks/scoring_example.ipynb - Tick “Activated” and save the changes.
-

- The next time the schedule pipeline runs, it will output the results to the Project Wiki
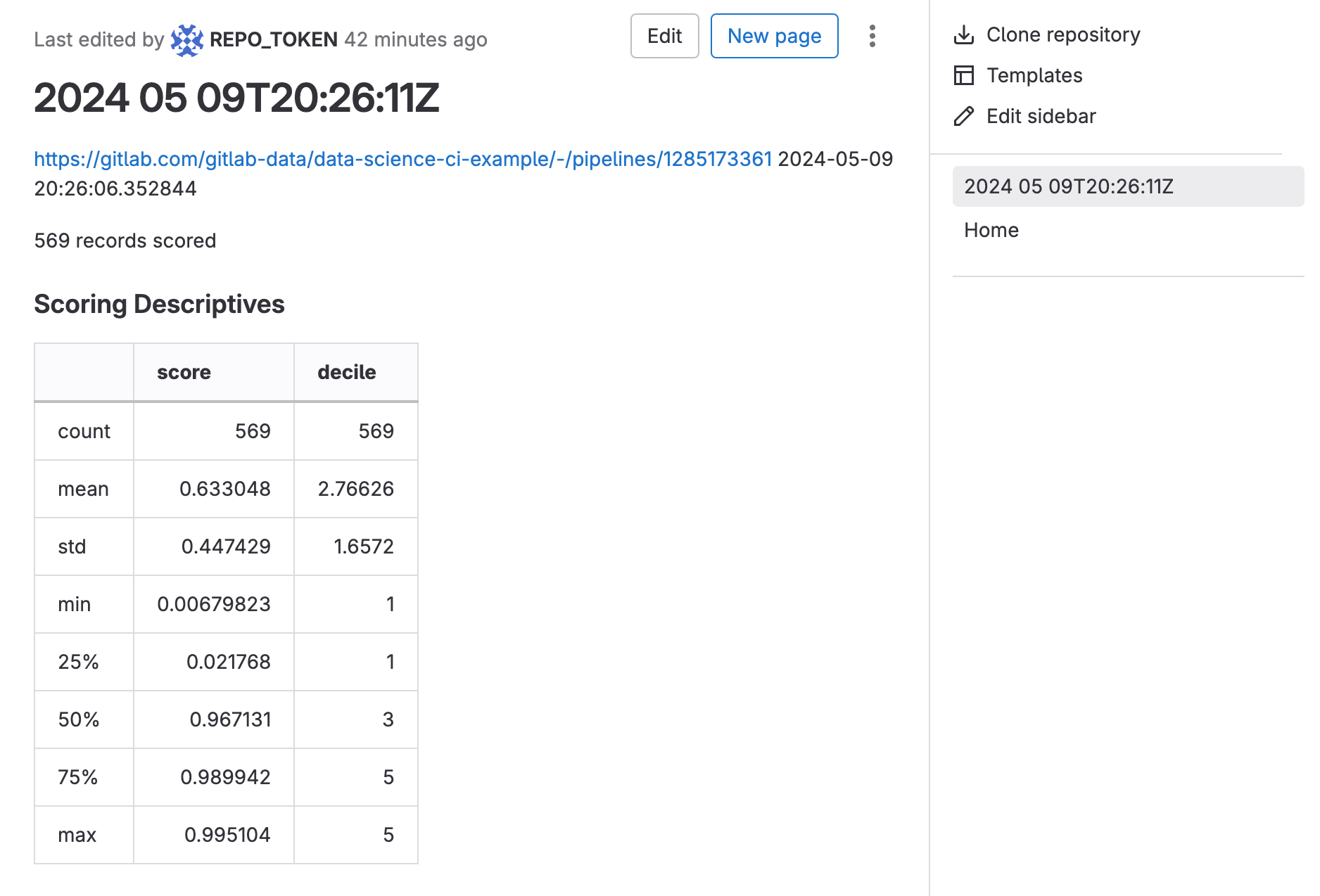
- Navigate to Plan -> Wiki and you will see a list by timestamp of all the times the scheduled pipeline has run, with links to the job logs and model metrics.
-
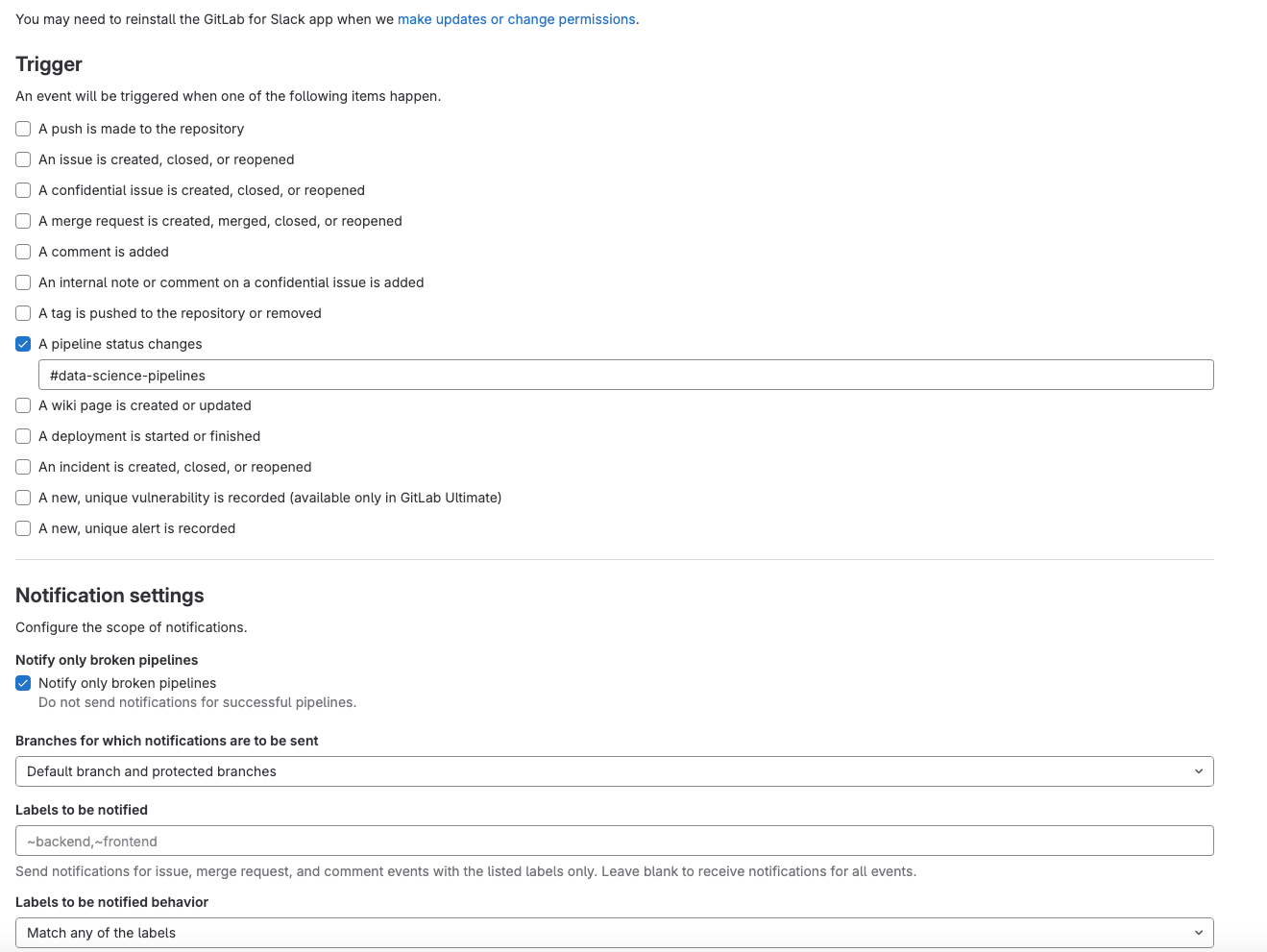
Slack Notifications (optional)
- In Slack, add the GitLab for Slack app
- Follow the instructions in the Gitlab For Slack app documentation
- We’ve setup our slack notifications so that notifications are sent to our #data-science-pipelines channel only when a pipeline fails. If a pipeline succeeds, a notification is not sent.
-
And that’s it! Feel free to modify these pipelines and notebooks to fit your data science modeling needs. And be sure to check out all the other great data science resources on our Data Science Handbook Page. If you are experiencing any difficulty or if you have any suggestions to improve these pipelines, feel free to open an issue with us. Happy pipelining!
46417d02)